Research Interests
We investigate the mechanisms underlying germline maintenance and accurate chromosome inheritance during meiosis. Addressing this is of vital importance in order to understand the sources of errors that result in infertility, miscarriages, birth defects such as Down syndrome, and tumorigenesis in humans. Our studies provide key insights into the molecular basis for the regulation of germline maintenance and meiotic chromosome segregation. Specifically, we apply combined genetic, molecular, cytological and biochemical approaches to:
- Understand the mechanisms promoting faithful meiotic chromosome inheritance at the molecular level and their regulation throughout meiotic progression.
- Explore meiotic chromosome dynamics and the roles of post-translational modifications in regulating chromosome synapsis, DNA double-strand break repair and accurate chromosome segregation.
- Investigate the roles played by regulators of the chromatin landscape, such as histone methyltransferases and demethylases, in germline maintenance and DNA double-strand break repair.
- Identify the meiotic pathways affected by exposure to environmental toxicants and develop high-throughput screening strategies for the identification of novel environmental meiotic disruptors.
Meiosis
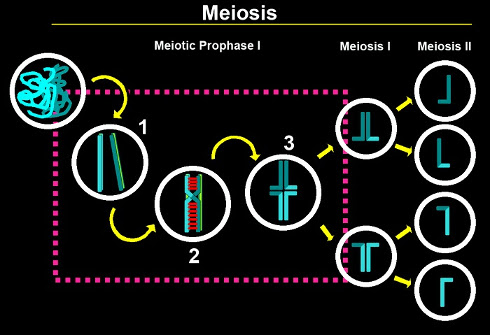
Meiosis is a specialized cell division program that results in the formation of haploid gametes from diploid germ cells. This cell division program is essential for the perpetuation of most sexually reproducing species and is critical for generating genetic diversity. The end result of meiosis (reduction of the chromosome complement by half) is accomplished by following a single round of DNA replication with two consecutive rounds of chromosome segregation (meiosis I and II). The partitioning of chromosomes needs to be tightly regulated to ensure that homologous chromosomes accurately segregate away from each other at meiosis I, and sister chromatids segregate away from each other during meiosis II. While meiosis II is similar to a mitotic division, chromosomes face a series of unique challenges during meiosis I. In order to overcome these challenges, chromosomes undergo a series of exquisitely orchestrated steps during meiotic prophase I: (1) pairing with their homologous partners, (2) formation of a "zipper-like" proteinaceous structure known as the synaptonemal complex (SC) between paired and aligned homologous chromosomes, and (3) completion of meiotic recombination leading to physical attachments (chiasmata) between homologs. Errors in pairing, synapsis or recombination ultimately lead to chromosome non-disjunction with disastrous consequences for the embryo.
Caenorhabditis elegans: a model system for studies of meiosis and germline maintenance.
We are addressing these aims in the nematode C. elegans. This is an extremely amenable model system for studies of germ cell maintenance and meiosis, which shares a high degree of conservation with humans. The germline accounts for more than half of the cells in the adult worm and its nuclei are distributed throughout the gonad in a defined order, correlating with the sequential stages of classical meiosis. Super- and high-resolution 3D imaging of meiotic chromosomes can be carried out in the context of a well preserved nuclear architecture, and pairing between homologs can be monitored by fluorescence in situ hybridization (FISH). Various approaches, including RNA-Seq and microarray analysis, applied to the C. elegans genome have led to the identification of meiotic gene candidates with germline-enriched expression. Techniques such as RNA-mediated interference (RNAi) and CRISPR-Cas9-guided genome editing allow for assessment of the function of germline-active genes.
We are applying a multipronged strategy to investigate the molecular basis of mechanisms underlying accurate meiotic chromosome segregation and meiotic chromosome dynamics. Here are some examples of the projects currently underway in the lab:
We are investigating the roles and the macromolecular assembly of the synaptonemal complex (SC), a structure at center stage during meiosis, whose functions are poorly understood and a matter of much debate despite its ubiquitous presence from yeast to mammals. The identification of six central region components of the SC in C. elegans (SYP-1, SYP-2, SYP-3, SYP-4, SYP-5 and SYP-6), coupled with well-established reagents and assays, allows us to address the following questions in the lab: What other components are required for synapsis? How are the assembly and disassembly of the SC regulated? Do the SYP proteins undergo specific post-translational regulation to promote changes in SC dynamics? How is the SC linked to progression of DSB formation and repair? What are the functions of this structure?
To address these questions we examined the interactions among the SC components by biochemical procedures and yeast two-hybrid analysis, and succeeded in investigating the organization of the SC by immuno-electron microscopy. These combined approaches led to the first model of the organization of the SC inC. elegans (Schild-Prüfert et al., 2011, Genetics). In addition, we are identifying new components and regulators required for synapsis via RNAi-based screens, yeast two-hybrid screens, and the identification of co-immunoprecipitated proteins. Ultimately, we will determine the role of both candidate SC components and their regulators in forming the SC structure, their effects on DSB formation, crossover recombination and meiotic progression.
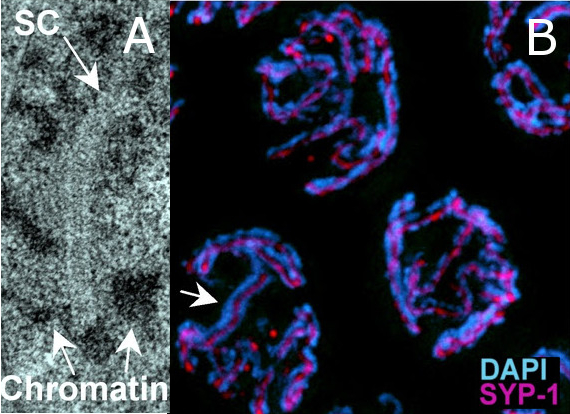
The SC visualized by EM analysis (A) and immunostaining (B) of pachytene nuclei in the C. elegans germline. (A) A continuous stretch of SC can be observed as a "zipper-like" structure flanked by electron-dense patches of chromatin. (B) Immunolocalization of SYP-1 (magenta) places this structural component of the SC at the interface between paired and aligned chromosomes (arrow indicates the SYP-1 signal flanked by the DAPI-stained chromosomes).
We have previously identified and characterized CRA-1, a conserved NatB domain-containing protein that uncouples chromosome synapsis from recombination, is required for SC formation (Smolikov et al., 2008, PLoS Genetics), and by antagonizing a previously unknown and conserved hydrolase ACER-1, controls the levels of acetyl-Coenzyme A and modulates global histone acetylation and meiotic DSB formation (Gao et al., 2015, PLoS Genetics). We also found that CRA-1 along with NATB-1 promote cotranslational N-terminal processing (N-terminal acetylation) of SYP-1, which is critical for SC assembly (Gao et al., 2016, Genes & Development). Our studies have also revealed that the dynamic process of SC disassembly occurs asymmetrically along the bivalents during late meiotic prophase, and correlates with the single crossover that forms towards the terminal thirds of chromosomes in C. elegans (Nabeshima et al., 2005, J Cell Biol). More recently, our studies of ECT-2, a homolog of mammalian Rho GEF, led us to find that SYP-2 phosphorylation is MPK-1-dependent and that the MAP kinase pathway coordinates crossover designation with the disassembly of SC proteins (Nadarajan et al., 2016, Elife). We also found that SYP-4 is phosphorylated in a Polo-like kinase (PLK-1/2)-dependent manner. SYP-4 phosphorylation depends on crossover designation, stabilizes the SC in pachytene by switching the central region of the SC from a more dynamic to a less dynamic state as shown by FRAP analysis, and negatively regulates DSB formation (Nadarajan et al., 2017, Elife). This identified a novel role for a central region component of the SC in the negative regulation of DSBs via a feedback loop triggered by crossover designation. We also discovered LAB-1, a novel protein we propose is the functional analog of Shugoshin, and showed that it is required for normal kinetics of SC disassembly and the protection of sister chromatid cohesion, both upon entrance into meiosis as well as in late meiotic prophase I, by targeting the protein phosphatase PP1 and restricting Aurora B kinase (Carvalho et al., 2008, Genes & Development; Tzur et al., 2012, PLoS Biology). Taken together, these studies have significantly contributed to our understanding of the process of SC assembly/disassembly, how changes in SC dynamics are linked to recombination and uncovered important new roles for the SC. Current and future studies in the lab are aimed at identifying additional components operating in these pathways, understanding the mechanism of function of these proteins, and investigating how chromosome remodeling during late prophase sets the stage for accurate chromosome segregation at meiosis I.
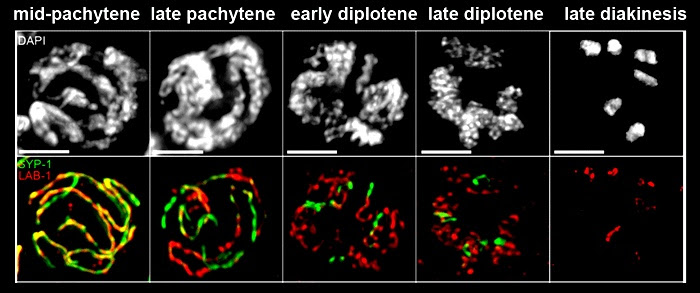
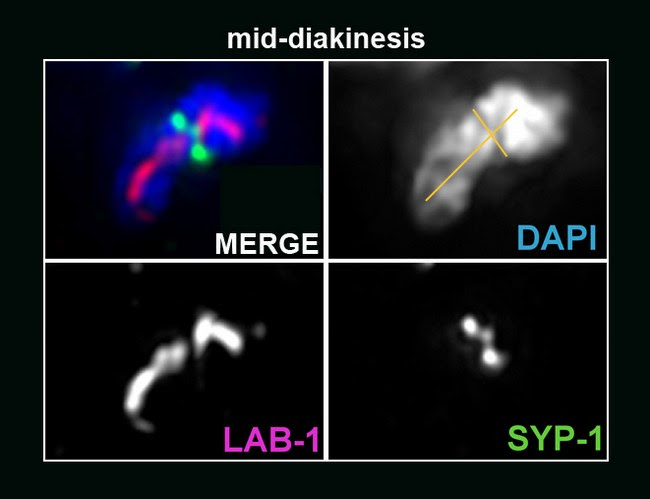
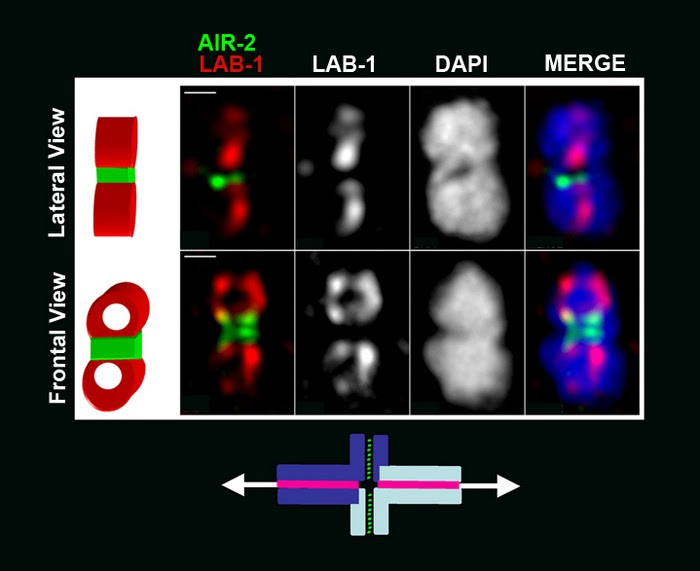
on the long arms whereas SYP-1 is present only on the short arms of the bivalent.
We are investigating the molecular basis for genomic stability and crossover formation in the germline. Through an RNAi screen for meiotic candidates we discovered HIM-18/SLX4 and ZTF-8/RHINO, conserved proteins required for the maintenance of genomic integrity. We showed that HIM-18/SLX4 acts as a scaffold promoting the processing of late homologous recombination intermediates (Saito et al., 2009, PLoS Genetics). Analysis of the conserved structure-specific endonucleases interacting with HIM-18/SLX4 showed that SLX-1 regulates crossover distribution along chromosomes (Saito et al., 2012, PLoS Genetics) and addressed the interplay between structure-specific endonucleases for crossover control during meiosis (Saito et al., 2013, PLoS Genetics). Our studies of ZTF-8/RHINO revealed it promotes repair at stalled replication forks and meiotic DSBs by transducing DNA damage checkpoint signaling via the 9-1-1 pathway (Kim and Colaiacovo, 2014, PLoS Genetics). Moreover, direct SUMOylation of ZTF-8 is required for its functions in DSB repair and DNA damage response (DDR), but not its localization (Kim and Colaiacovo, 2015, Genetics), providing new insights into the post-translational regulation of this conserved DDR and DSB repair factor. Future studies are aimed at investigating the roles played by other proteins we are discovering act in promoting genomic stability throughout the germline.
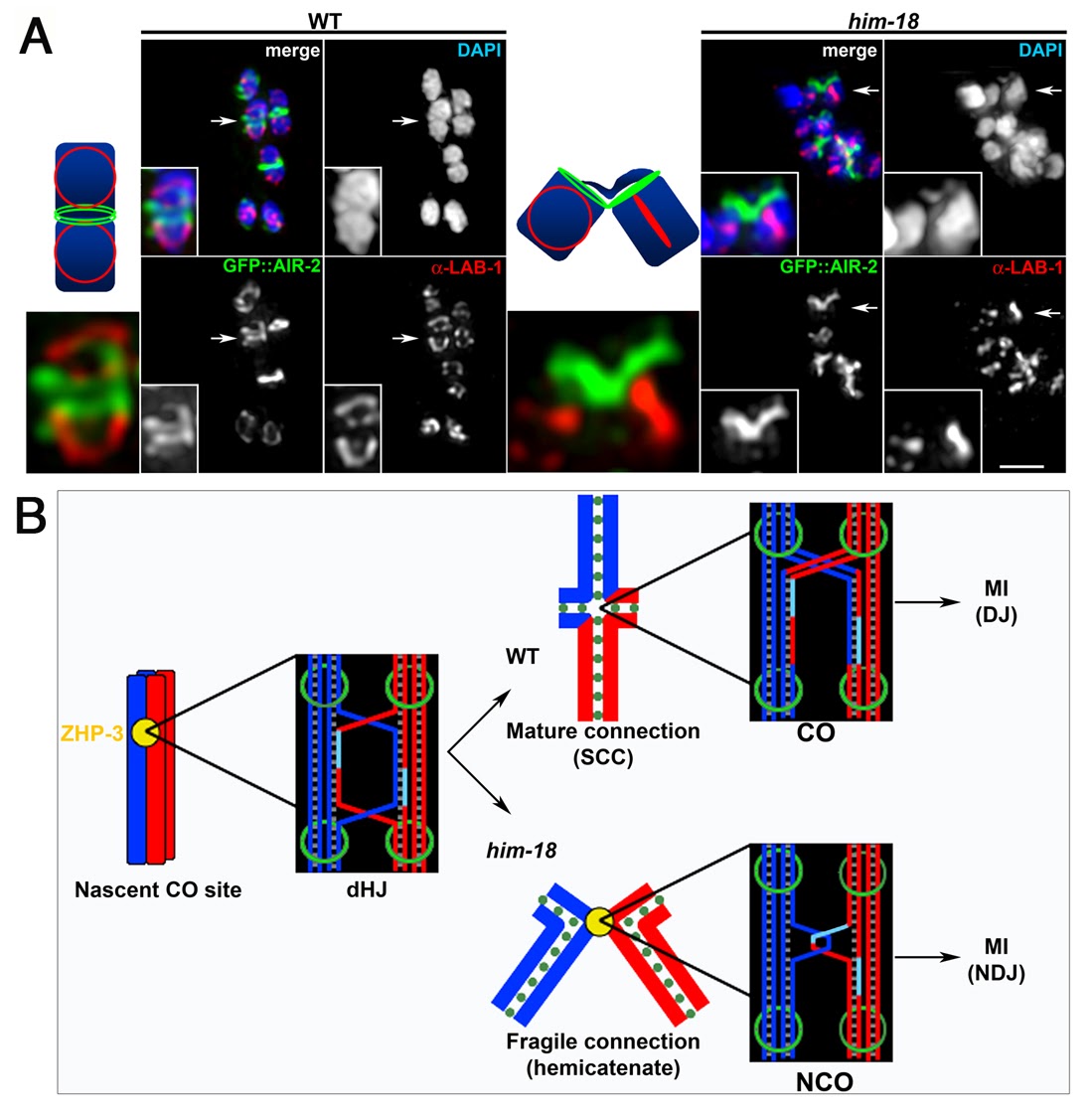
HIM-18/Slx4 is required for bivalent stability. (A) Bivalents in prometaphase I oocytes in wild type and him-18 mutants were examined for LAB-1 and GFP::AIR-2 localization to highlight both axes of this configuration. Arrows indicate the bivalents enlarged in the insets. The illustrations depict the chromosome axis configurations. Bar, 2 µm (B) Meiotic crossover formation. Paternal sister chromatids are blue and maternal sister chromatids are red. ZHP-3 marks the nascent CO site during late pachytene. In wild type, HIM-18-dependent dHJ processing results in the formation of a chiasma. Sister chromatid cohesion (SCC) contributes to the tension promoting the proper alignment of the bivalent at the metaphase I plate. Disjunction (DJ) of the homologous chromosomes following degradation of SCC along the mid-bivalent region occurs at anaphase I (MI). In him-18 mutants, dHJ processing is impaired. Dissolution intermediates such as hemicatenane structures or the complete dissolution of the dHJ may manifest in chromosome bridges and lagging chromosomes observed at anaphase I resulting in chromosome non-disjunction (NDJ). Green circles and rings represent cohesin.
Chromatin structure plays critical roles in most chromosome functions in vivo, including DNA replication, chromosome segregation, and transcription. The roles of histones, the main protein components of chromatin, are controlled to a significant degree by post-translational modifications along their N-terminal tails. One such critical type of modification is histone methylation, which has been implicated in biological processes such as the establishment and maintenance of heterochromatin, transcriptional regulation, X inactivation and DNA damage response. A search for histone tridemethylases in the laboratory of our collaborator, Dr. Yang Shi (Dept. of Pathology, HMS), led to the identification of JMJD2A. This protein is also present in C. elegans and we proceeded to investigate its in vivo biological relevance by focusing on the roles played by CeJMJD2 in the germline. We determined that depletion of this candidate by RNAi leads to an increase in H3-K9Me3 and H3-K36Me3 levels on chromosomes in the adult hermaphrodite germline. This was accompanied by a transient increase in RAD-51 foci and a p53-dependent increase in germ cell apoptosis indicating the activation of a DNA damage checkpoint (Whetstine et al., 2006, Cell). These studies were the first demonstration of the existence of histone demethylases able to reverse lysine trimethylation.
Our subsequent analysis of SPR-5, a homolog of the lysine specific demethylase LSD1, revealed that it shows enzymatic activity towards H3K4me2 both in vitro and in vivo and plays a role in meiotic DSB repair in C. elegans (Nottke et al., 2011, PNAS). We are currently applying combined genetic, molecular, biochemical and cytological approaches to investigate the roles of SPR-5, as well as related lysine specific demethylases and other chromatin regulators, in DNA damage repair and germline maintenance.
Despite the severe outcomes of impaired meiosis, the screening and analysis of environmental toxicants for their ability to disrupt this process have been particularly challenging. Meiosis is a complex cellular program, which in the case of mammalian female meiosis spans from several months in mice to several decades in humans. It is initiated early on during embryogenesis, where key events guiding chromosomal segregation and exchange take place, and is only completed at puberty during ovulation. Each of these crucial steps of meiotic prophase I, namely chromosome pairing, synapsis and recombination, occur at specific stages of development within the confines of the embryonic gonad and are therefore not easily accessible for study. Due to time, cost and experimental constraints, the study of environmental effects on the process of meiosis in mammals is therefore highly challenging.
Our goal is to improve our ability to efficiently and comprehensively interrogate our chemical environment for its effects on reproduction and meiosis. Therefore, we have set out to establish the nematode C. elegans as a fast and reliable meiotic toxicological model relevant for mammalian meiosis. Specifically, our aim is to improve the safety of toxicological testing by including, for the first time, a strategy that addresses the effect of compounds and mixtures on reproduction via alteration of the meiotic program in a multicellular organism.
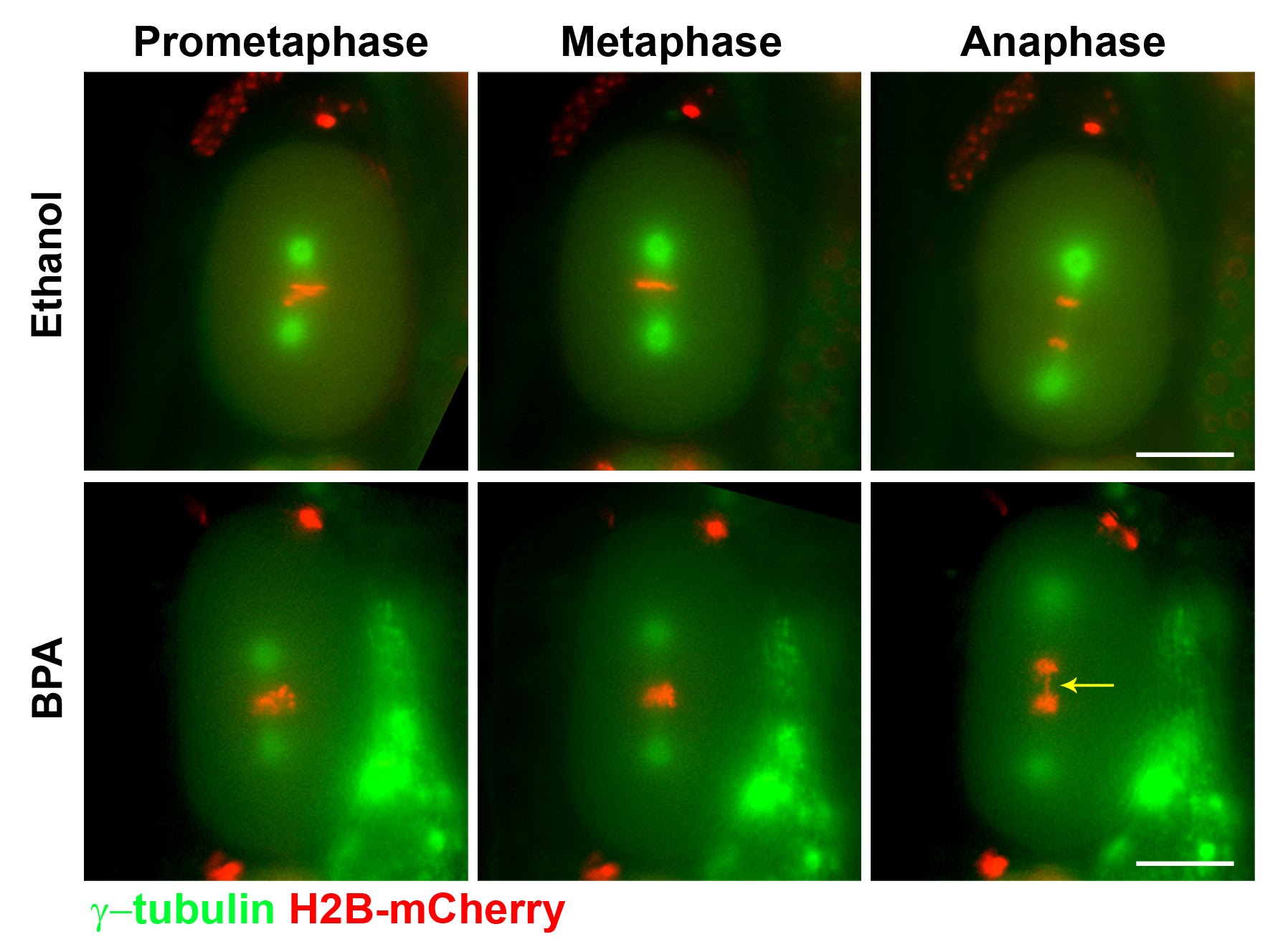
As a “proof-of-concept” we investigated the effects of exposure to Bisphenol A (BPA), a highly prevalent constituent of plastics that has been associated with diabetes, cardiovascular disease and an increased risk of miscarriages in humans. In mice, BPA exposure disrupts the process of meiosis, however, analysis of the affected molecular pathways is lagging and has been particularly challenging. We showed that exposure of the nematode Caenorhabditis elegans to BPA, at internal concentrations consistent with mammalian models, causes increased sterility and embryonic lethality (Pallard and Colaiacovo, 2010, PNAS; Pallard and Colaiacovo, 2011, Cell Cycle). BPA exposure resulted in impaired chromosome synapsis and disruption of meiotic double-strand break repair (DSBR) progression. BPA carries an anti-estrogenic activity in the germline and resulted in germline-specific down-regulation of DSBR genes thereby impairing maintenance of genomic integrity during meiosis. Follow up studies revealed that BPA exposure induces oxidative DNA damage in the germline that can be rescued with the antioxidant Coenzyme Q10 (CoQ10), which neutralizes DNA damage resulting from oxidative stress and could constitute a low-risk and low-cost strategy to attenuate the impact of BPA on fertility (Hornos Carneiro et al., 2020, Genetics). C. elegans therefore constitutes a model of remarkable relevance to mammals with which to assess how our chemical landscape affects germ cells and meiosis.
Subsequent studies in the lab established a high-throughput screening platform using C. elegans to identify environmental toxicants inducing aneuploidy as a result of impaired germline function (Allard et al., 2013, Environ Health Perspect; Shin et al., 2019, PLoS Genetics). This strategy was applied to the analysis of chemicals that are widely present in our environment including pesticides, phthalates, and chemicals used in hydraulic fracturing and crude oil processing (Shin et al., 2019, PLoS Genetics). Analysis of exposures to dibutyl phthalate (DBP), and the pesticides TCMTB, and permethrin (Shin et al., 2019, PLoS Gen), as well as diethylhexyl phthalate (DEHP) (Cuenca et al., 2020, PLoS Genetics) revealed mechanisms by which these chemicals are affecting reproductive health. Current work is focused on understanding how exposure to pesticides and phthalates affects the germline and their sexually dimorphic effects.